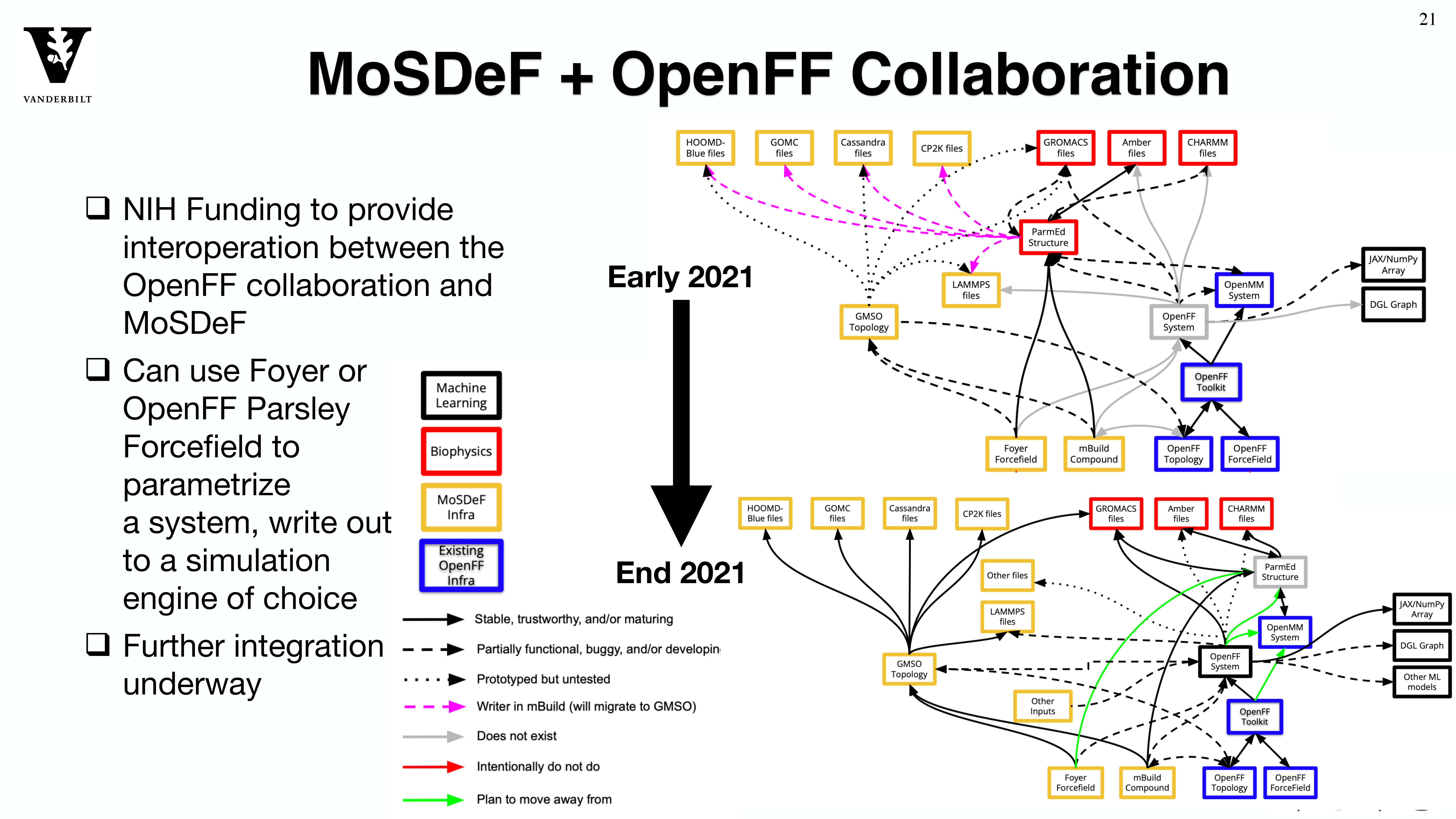
TRUE Simulations
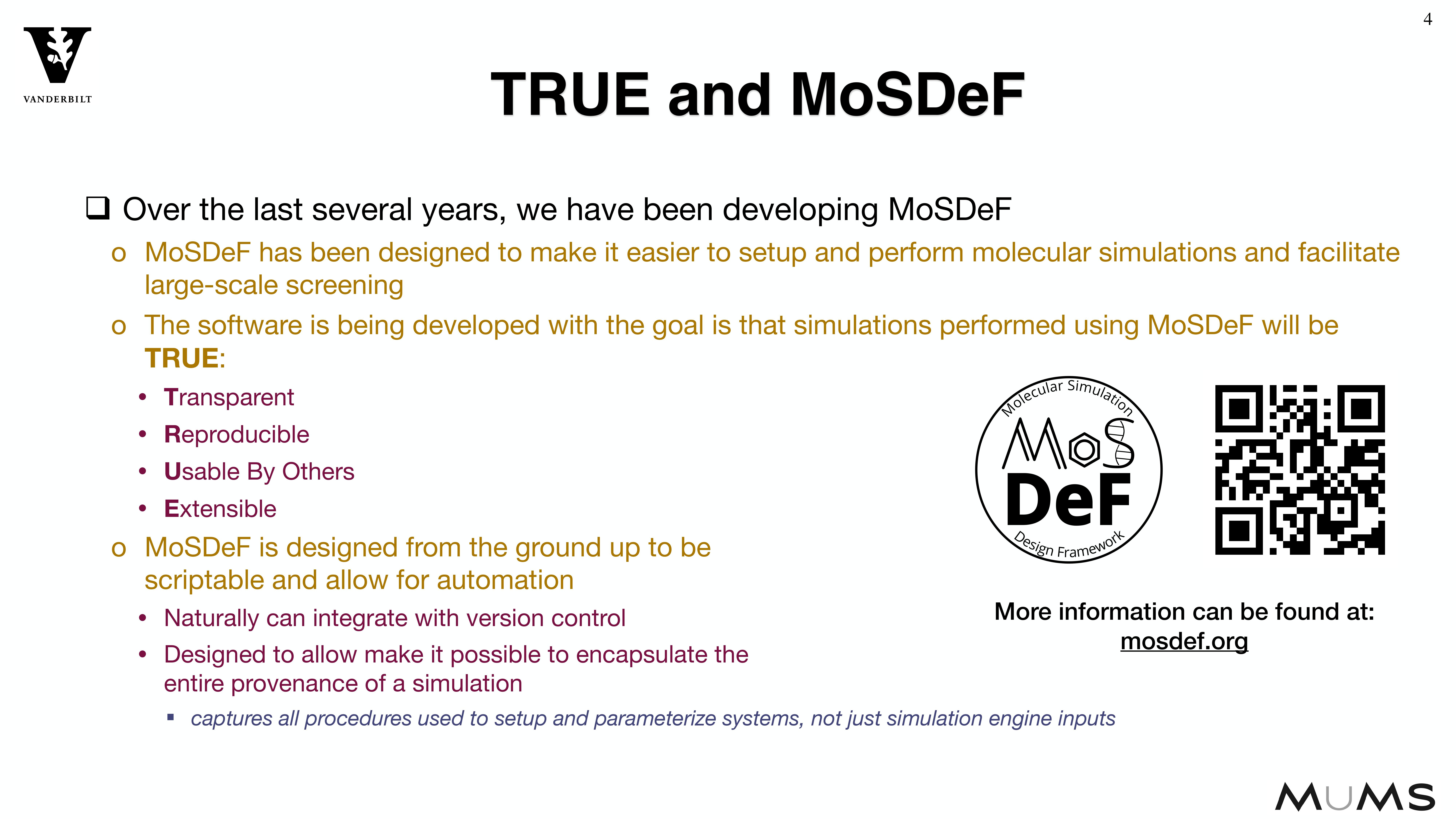
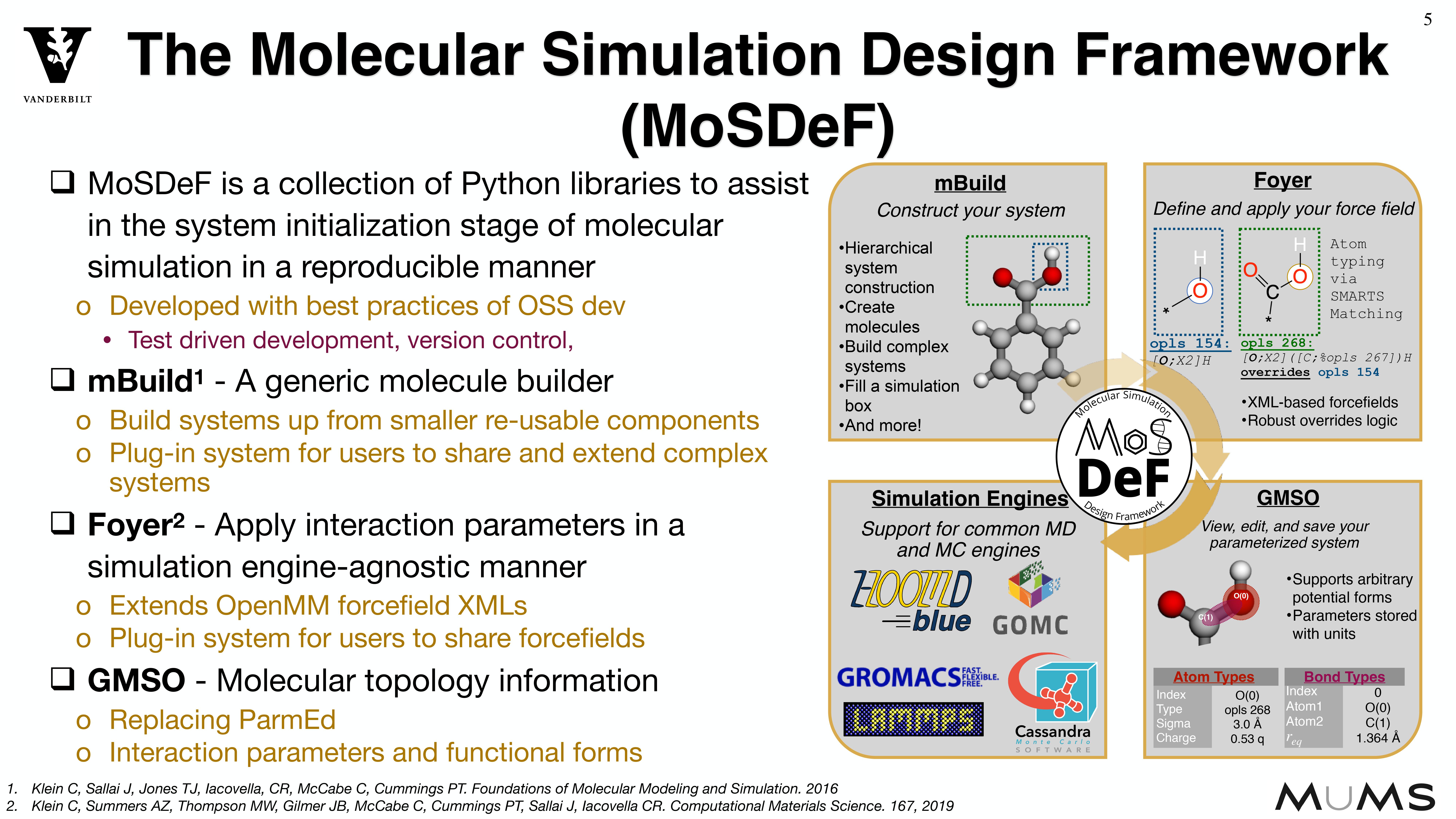
Through the MoSDeF integrated framework, the exact procedures used to set up and perform simulation workflows and associated metadata (i.e. the provenance) can be scripted, encapsulated, version-controlled, preserved, and later reproduced by other researchers. This allows molecular simulation studies to be conducted and published in a manner that is TRUE : “Transparent, Reproducible, Usable by others, and Extensible .” The following paper highlights these principles in details.

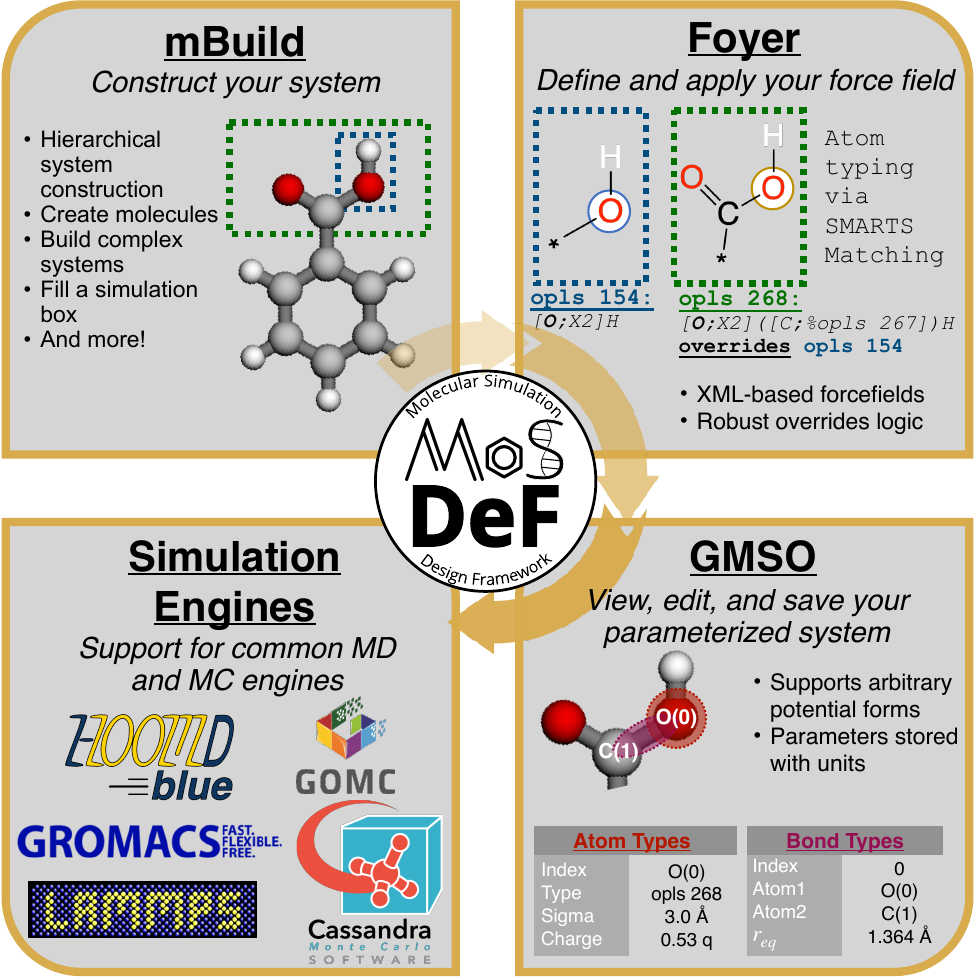
Flexibility
System construction and initialization are performed in a simulation engine agnostic fashion,
allowing users to use a single set of scripts to generate inputs for a variety of different
simulation engines.
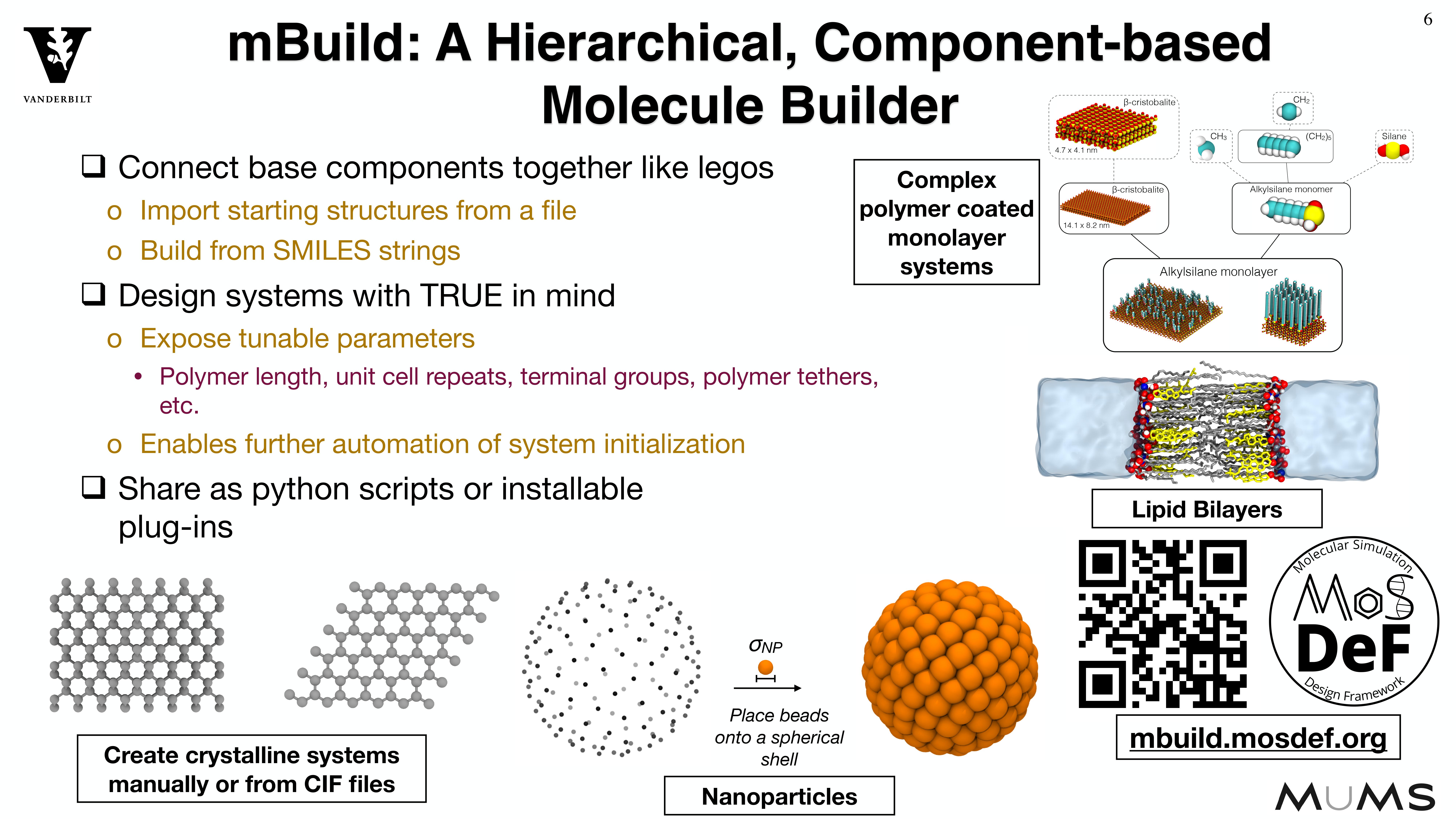
Because systems are constructed in a hierarchical, scriptable fashion, modifications to system
chemistry can be easily made, allowing for screening over chemical space.
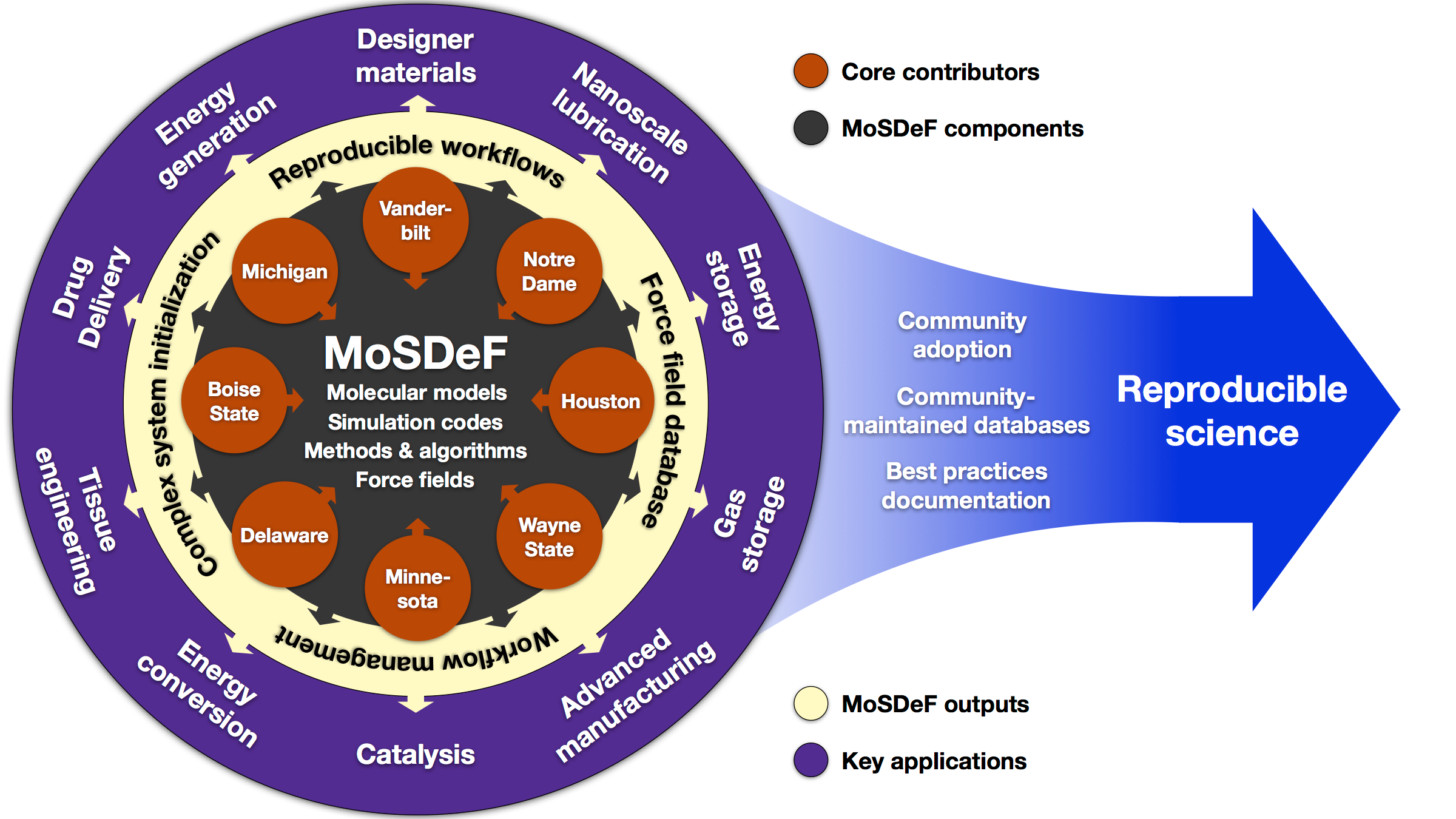
Community
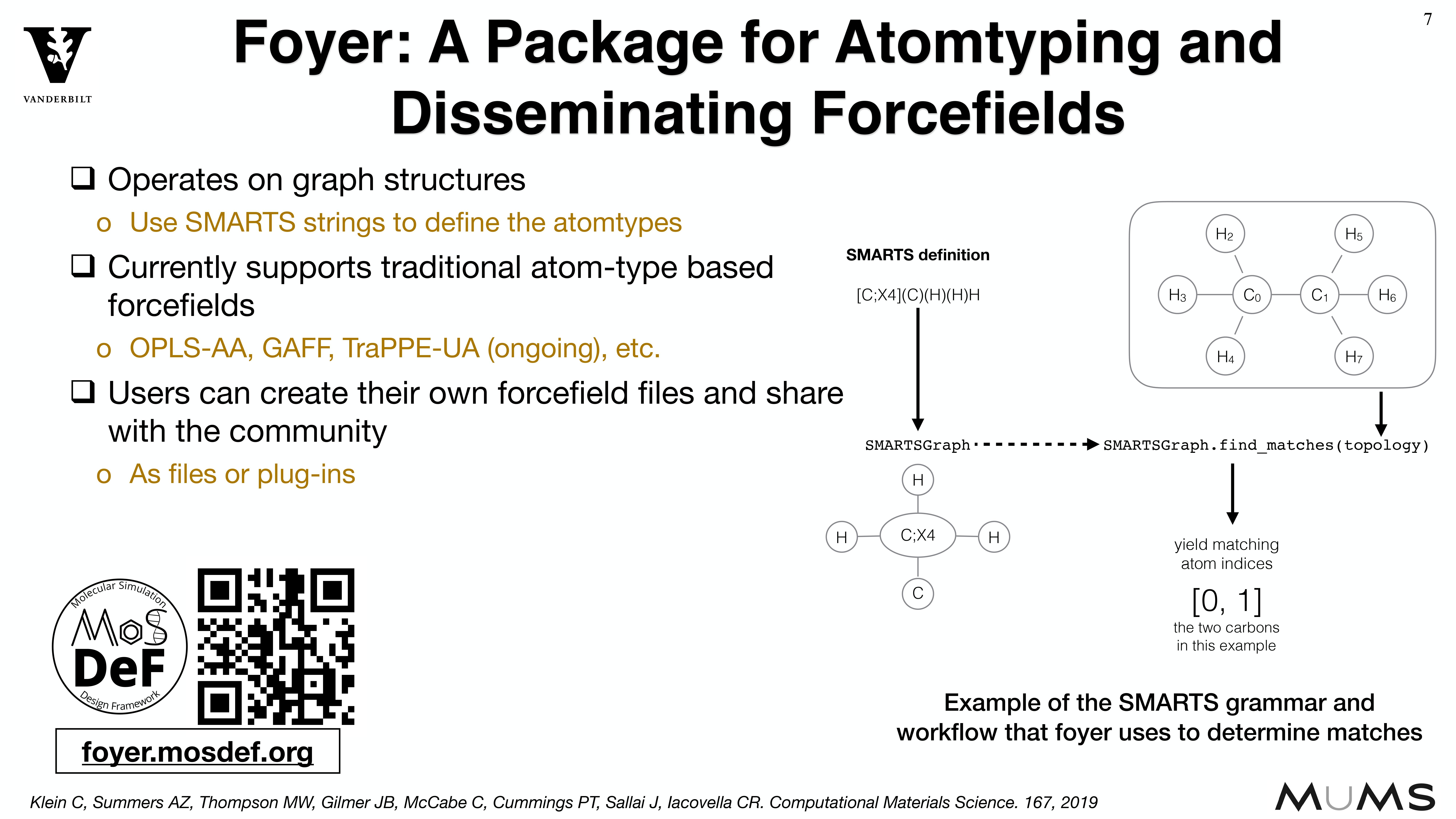
MoSDeF is designed to allow system construction scripts to be easily made into easy to distribute libraries; similarly, forcefield files contain parameters and their usage rules that can be easily shared and version controlled.
Join the mailing list
Mosdef Gitter
Github Organization

Quick Start
| Details | GitHub | Binder |
|---|---|---|
| MosDeF Tutorials: Curated tutorials for the MosDeF Framework |
|
|
| mBuild Tutorials: Curated tutorials for mBuild |
|
|
| Foyer Tutorials: Curated tutorials for Foyer |
|
Quick Peek
Foundational MoSDeF Repositories
Other MoSDeF Projects
Projects Using MoSDeF
Also, checkout projects from our collaborators: Boise State | Michigan | Notre Dame | Wayne State.
Collaborators
MoSDeF is supported by a Cyberinfrastructure for Sustained Scientific
Innovation (CSSI) grant funded by the National Science Foundation Office of Advanced
Cyberinfrastructure.
Get Involved
Chat
Having issues with any of the packages or want to contribute to the project? Join us on gitter.
ChatCheck out our recent news
We are constantly making new releases. Please check here for release announcements.
AnnouncementsContribute
Contributions are welcome! Just fork any of the projects and submit some pull requests or develop your own custom extensions.
Contribute